Introduction
Nuchal translucency (NT) is a subcutaneous fluid collection behind the fetal neck that is physiologically present in the first trimester in all fetuses. This measurement is well standardized and so measured by ultrasound between 11+0 and 13+6 weeks of gestation. The association between increased NT and chromosomal abnormalities has been described since 1992 [1-3]. It is current knowledge in fetal medicine that NT thickening is associated not only with chromosomal aneuploidies but also with genetic syndromes, fetal congenital malformations and increased risk of miscarriage or intrauterine demise [4-7]. NT is determined as an absolute value or according to percentile curves based on crown-rump length (CRL) of the fetus (45-84 mm). A nuchal measurement ≥ 3.5 mm or ≥ 99th percentile is a well-known indication for invasive testing by amniocentesis or chorionic villus sampling (CVS) [8-10]. However, several studies have demonstrated an increased risk of chromosomal abnormalities for fetuses with a NT between the 95th and 99th percentiles [11-14]. Nevertheless, there are no uniform guidelines regarding whether to perform an invasive test in this subgroup.
Given the current use of genome-wide cfDNA testing in our center as routine screening, the objective of this study was to identify chromosomal abnormalities detected in the subgroup of NT between the 95th-99th percentiles and to evaluate the accuracy of genome-wide cfDNA testing for their detection.
Methods
This single-center retrospective study, conducted at Centre Hospitalier Universitaire (CHU) of Liège, included all pregnancies with increased NT ≥ 95th percentile on the first-trimester ultrasound between January 2017 and December 2023. At the first-trimester morphological ultrasound, if the NT is below the 95th percentile, the patient is offered a non-invasive prenatal testing (NIPT) by a cfDNA test . If the NT measurement is above the 95th percentile, we propose all patients to undergo invasive testing via amniocentesis or CVS for chromosomal microarray analysis (CMA) and genetic counseling. All genetic analyses were performed in the clinical genetics laboratory of the CHU de Liège. During this period, 59 samplings were performed solely based on NT ≥ 95th percentile. The cohort was stratified into two groups based on NT measurement: NT ≥ 99th percentile, corresponding to 3.5 mm (Group A), and NT between 95th-99th percentile (Group B). Group B was further divided into two subgroups: NT between 3-3.4 mm (Group B1) and NT ≤ 2.9 mm (Group B2). The results obtained by CMA were then compared to the results that would have been obtained using genome-wide cfDNA testing. Our NIPT is a lab-developed (LDT) whole genome next-generation sequencing (NGS) method. NGS is performed using a NextSeq550 sequencer (Illumina). Genome-wide genomic representation profiling and interpretation (pipeline version 4.6) is performed as described by Bayindir et al. (2015) and detects classical trisomies (T13-T18-T21), as well as other autosomal aneuploidies. Regarding copy number variations (CNVs), our NIPT has been confirmed to detect maternal duplications or deletions of approximately 0.2 Mb (200 kb) or more. The cut-off applied by the clinical genetics laboratory to report fetal CNVs is 20 Mb (20 000 kb) although it is possible to detect smaller variants. For the purposes of this paper, we assumed that NIPT would detect 100% of aneuploidies.
Ethics approval
Ethics approvals were obtained from the CHU of Liège of research ethics committee.
Results
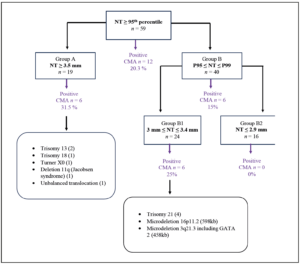
CMA and NT results were obtained for 59 patients with NT measurements including and above the 95th percentile. Group A (NT ≥ 3.5 mm) consisted of 19 fetuses, while Group B (NT 95th – 99th percentile) comprised 40 fetuses. Group B1(NT 3.0-3.4 mm) included 24 fetuses, and Group B2 (NT ≤ 2.9 mm) included 16 fetuses. In the total group (n=59), 12 chromosomal abnormalities were identified via CMA, representing 20.3% of cases. Among those in Group A (n=19), 6 abnormalities were observed, accounting for 31.5%. In Group B, 6 abnormalities were detected, representing 15%. It is noteworthy that all abnormalities found in Group B fell within Group B1, and no anomalies were detected in Group B2, corresponding to NT values below or equal to 2.9 mm (Figure 1).
The abnormalities found in the various groups are depicted in Figure 1. Within Group B, more specifically Group B1, there were 4 cases of T21 and 2 CNVs, specifically 2 microdeletions measuring
0.598 Mb and 0.458 Mb respectively. The first deletion affects the 16p11.2 region, encompassing 19 genes located in the syndromic 16p11.2 region. The other deletion affects the 2q21.3 chromosomal region, encompassing 14 genes and including the entire GATA2 gene.
Based on these results of Group B, we can theoretically deduce that the 4 aneuploidies (T21) would have been detected even by a genome-wide and a targeted cfDNA testing. Regarding the two microdeletions, their sizes fall significantly below the resolution of fetal CNV detected by our cfDNA testing and would have been detected only if inherited from the mother. The deletion 16p11.2 was a
CNV inherited from the mother and the cfDNA testing could have detected a maternal CNV in this case (with a resolution of 0.200 Mb). However, the other microdeletion was not found in either parent, indicating a de novo CNV that would not have been detected with cfDNA testing because it was under the resolution cut-off.
Discussion
Several previous studies have indicated that fetuses with an NT measurement between the 95th and 99th percentiles have an elevated risk of chromosomal abnormalities [12-14]. However, there is no clear international consensus on screening for this abnormal NT subgroup. The definition of an increased NT varies among different sources. Some authors define it as a measurement ≥ 3.5 mm or
≥ the 99th percentile, while others suggest lower thresholds for considering genetic diagnosis, such as 3.0 mm or 95th percentile. The International Society of Ultrasound in Obstetrics and Gynecology (ISUOG) does not provide a specific definition for "increased NT" [15], but the American College of Obstetricians and Gynecologists (ACOG) and the Society for Maternal-Fetal Medicine (SMFM) now recommend thresholds of 3.0 mm or the 99th percentile, although it's important to note that these two definitions are not equivalent [16].
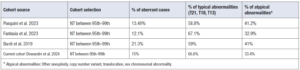
In our center, the cut-off for performing an invasive test is the 95th percentile. Our results demonstrate that the overall detection rate of a chromosomal abnormality in fetuses with NT between the 95th-99th percentile (Group B) is 15%. Despite our small cohort, these data are similar to those from larger-scale studies. In the study of Pasquini et al. [12], the incidence of chromosomal abnormalities was 13.49% for NT between the 95th and 99th percentile. Fantasia et al. [13] reported an incidence of chromosomal abnormalities of 12.1% for this intermediate NT subgroup. In a study by Bardi et al. [14] the incidence was 20.3%. In this group of fetuses, the most frequently encountered chromosomal abnormalities are common trisomies, with trisomy 21 being the most frequent, accounting for approximately 50-60% of cases. Other frequently encountered abnormalities include CNVs. A significant portion of samples with abnormal results represents chromosomal mosaicism. As for atypical anomalies, one of the most common is the deletion of 22q11.2, corresponding to DiGeorge Syndrome (Table 1).
Note that in our study, all patients underwent an invasive procedure using CMA as a first-line diagnostic method. In the published studies, the invasive test was only proposed to patients who had a high risk on combined screening (NT, age, and serum markers). Moreover, some studies use conventional karyotyping as the first-line approach instead of CMA.
In several countries including Belgium, CMA has replaced classic karyotyping (with a resolution between 5 and 10 Mb) following invasive sampling, and CMA is currently the most effective diagnostic test in prenatal diagnosis [17,18]. The current consensus size cut-off for CNVs in the prenatal context ranges from 0.2 Mb to 0.4 Mb, aiming to minimize the impact of variants of uncertain significance (VUS) [19,20].
Regarding non-invasive prenatal testing, genome-wide cfDNA testing has transformed prenatal care since its introduction. In 2017, Belgium became the first country to fully implement and reimburse cfDNA testing as the primary screening test for all pregnant women [21]. In addition to cfDNA screening for common trisomies (targeted cfDNA testing), some companies have added other aneuploidies and CNVs to their screening panels. Some have limited their approach to specific microdeletions, while others report a genome-wide approach detecting gains and losses ≥ 7 Mb [17,22]. All academic genetic centers in Belgium use a genome-wide cfDNA testing that detects all aneuploidies and fetal CNVs with a resolution of 20 Mb.
In our study, assuming cfDNA testing to have a 100% detection rate for aneuploidies, the 4 trisomies would have been detected by genome-wide cfDNA testing. Regarding the 2 CNVs, the deletion of the 16p11.2 region inherited from the mother would have been detected as a maternal CNV. The de novo deletion including the GATA2 gene would have been missed by NIPT screening. An anomaly of the GATA2 gene predisposes to myeloid malignancy and immunodeficiency including recurrent infections, lymphedema and sometimes urogenital malformations. In this pregnancy, no structural abnormality was diagnosed during follow-up ultrasound, indicating that this anomaly would not have been detected later during the pregnancy. The two microdeletions did not result in a request for pregnancy termination.
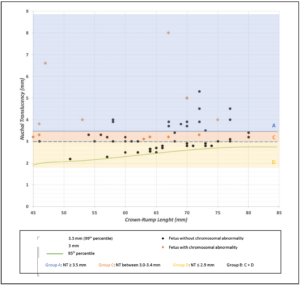
Finally, in our group of fetuses with an NT measurement between the 95th and 99th percentiles, we observed that all anomalies were detected in Group B1 (NT 3-3.4 mm) and none were detected in Group B2 (NT ≤ 2.9 mm). The proposed cut-off for invasive procedures could be 3 mm instead of the 95th percentile. According to a study by Hui et al. [16] there is a 0.2% prevalence of chromosomal abnormalities in fetuses with NT < 3 mm. Another study by Maya et al. [23] also proposes that the NT cut-off for invasive testing should be 3.0 mm. They describe a rate of 1.7% of chromosomal anomalies in fetuses with an NT ≤ 2.9 mm. Our findings align with these studies, as we observed a 0% rate of chromosomal or genetic anomalies in this subgroup, although this rate may be slightly underestimated due to our smaller cohort size (Figure 2). Using 3 mm as cut-off instead of the 95th percentile, the sensitivity for detecting chromosomal abnormalities will decrease while the specificity will increase [11].
In agreement with our results, and the increasing evidence of a very low risk of procedure-related miscarriage after an invasive procedure [24-26], CMA remains the procedure that offers the best detection rate and is the most comprehensive analysis when women wish to be informed of as many chromosomal aberrations as possible. However, genome-wide cfDNA testing can be an alternative for patients who do not wish to undergo an invasive procedure. In cases where NIPT is chosen as the initial screening method, patients must be informed that it is a screening test associated with a residual risk. Our cohort is too small to quantify this residual risk; larger-scale studies are necessary.
It is important to inform the patient that any abnormal NIPT results necessitate further confirmatory diagnostic testing through an invasive procedure. This sometimes leads to a prolonged period of anxiety while awaiting definitive results. Timely provision of comprehensive and accurate information during early gestation is crucial for enabling informed decision-making by patients.
Therefore, genetic counseling before and after testing is essential [27].
Conclusion
Our study shows that fetuses with mildly increased NT (95th-99th percentiles) have a 15% risk to be affected by a chromosomal aberration. Based on our results, we suggest that invasive procedures by CMA offer the best detection rate and are indicated in this population, using a cut-off of ≥ 3 mm.
Nevertheless, if a pregnant woman declines an invasive procedure, then genome-wide cfDNA testing can be proposed as a good alternative to invasive test. In this case, the patient should be informed that there is a residual risk for a fetal chromosomal abnormality even in the presence of a normal genome-wide test.
Strengths and limitations
The strength of our study is that we included all cases of fetuses with an NT between the 95th-99th percentile irrespective of a combined test risk. Moreover, all patients underwent fetal CMA. The routine use of genome-wide cfDNA testing instead of targeted testing for trisomies adds value to our study. The main limitation of our study is our relatively small number of patients,
although our study is representative of larger studies. Another potential limitation is the measurement of NT by various sonographers, with no available data on possible interobserver variation. Although most of the ultrasounds were performed by certified sonographers from our center, some patients were referred from external centers. In these centers, not all sonographers are certified, which is why we cannot formally confirm that there is no overestimation or underestimation of some nuchal measurements.
Conflict of interest statement
The authors have no conflicts of interest to declare.